Fine-tuning a Transformers model with medkit#
Note
This example may require optional modules from medkit, use the following to install them:
pip install 'medkit-lib[training,hf-entity-matcher]'
In recent years, Large Language Models (LLMs) have achieved very good performance in natural language processing (NLP) tasks. However, training a LLM (involving billions of parameters) from scratch requires a lot of resources and large quantities of text.
Since these models are trained on general domain data, they learn complex patterns. We can adapt (fine-tune) the last layers to a specific task using our data and low resources. LLMs are PreTrained and accessible with libraries like 🤗 Transformers. Medkit has some components to fine-tune these models.
Loading the QUAERO dataset#
To fine-tune a model, we need data. In this tutorial, we will use the QUAERO dataset (https://quaerofrenchmed.limsi.fr/). The QUAERO dataset is a mix of biomedical article titles and medication leaflets, in french. Entities were annotated with UMLS semantic groups labels (ex: “ANAT”, “CHEMI”, “DISO”, “PROC”, etc). The dataset is available in the BRAT format. Let’s download it:
import os
import urllib
import zipfile
QUAERO_URL = "https://quaerofrenchmed.limsi.fr/QUAERO_FrenchMed_brat.zip"
QUAERO_DIR = "QUAERO_FrenchMed/corpus/"
# if not os.path.exists(QUAERO_DIR):
# !wget -O quaero.zip https://quaerofrenchmed.limsi.fr/QUAERO_FrenchMed_brat.zip
# !unzip -o ./quaero.zip
The corpus has been pre-split into train/dev/test groups, and inside each split the documents are grouped between EMEA files (drug leaflets) and MEDLINES files (article titles):
QUAERO_FrenchMed/corpus
├── train
│ ├── EMEA
│ │ ├── 118_1.txt
│ │ ├── 118_1.ann
│ │ ├── ...
│ ├── MEDLINE
│ │ ├── 128732.txt
│ │ ├── 128732.ann
│ │ ├── ...
├── dev
│ ├── EMEA
│ ├── MEDLINE
├── test
│ ├── EMEA
│ ├── MEDLINE
QUAERO was annoated with overlapping entities, but the NER operation we will
fine-tune only supports non-overlapping entities, so we want to get rid of any
overlaps in our training data by keeping only the longest entity when 2 entities
overlap. medkit provides a
filter_overlapping_entities() helper function for this
purpose.
Instead of directly loading all the documents and annotations in each subdirectory with
load(), we will load each file separately with
load_annotations() and filter the annotations with
filter_overlapping_entities():
from glob import glob
from medkit.core.text import TextDocument
from medkit.io.brat import BratInputConverter
from medkit.text.postprocessing import filter_overlapping_entities
converter = BratInputConverter()
# Load quaero docs without overlapping entities
def load_quaero_docs(quaero_dir):
docs = []
for text_file in sorted(glob(quaero_dir + "*.txt")):
# Load the .txt and create an empty doc
doc = TextDocument.from_file(text_file)
# Load the entities from the correspoding .ann file
ann_file = text_file.replace(".txt", ".ann")
entities = converter.load_annotations(ann_file)
# Remove overlapping entities (by keeping the longest)
entities = filter_overlapping_entities(entities)
# Add the filtered entities to the document
for entity in entities:
doc.anns.add(entity)
docs.append(doc)
return docs
# To this for each split, both for EMEA and MEDLINE files
train_docs_emea = load_quaero_docs(QUAERO_DIR + "train/EMEA/")
train_docs_medline = load_quaero_docs(QUAERO_DIR + "train/MEDLINE/")
dev_docs_emea = load_quaero_docs(QUAERO_DIR + "dev/EMEA/")
dev_docs_medline = load_quaero_docs(QUAERO_DIR + "dev/MEDLINE/")
test_docs_emea = load_quaero_docs(QUAERO_DIR + "test/EMEA/")
test_docs_medline = load_quaero_docs(QUAERO_DIR + "test/MEDLINE/")
Splitting QUAERO EMEA docs in sentences “mini-docs”#
While the MEDLINE documents are very short because each doc is just an article title, the EMEA documents are longer and have to be split by sentence in order to be usable during training. Here is the code to split each EMEA doc by sentence and create new mini-docs for each sentence with the entities they contain:
from medkit.core.text import Entity, Span
from medkit.text.segmentation import SentenceTokenizer
from medkit.text.postprocessing import DocumentSplitter
# Split an EMEA document into multiples "mini-docs",
# one for each sentence
def split_emea_docs(docs):
sentence_tokenizer = SentenceTokenizer(
output_label="sentence",
# EMEA docs contain one sentence per line so splitting on newlines should be enough
split_on_newlines=True,
punct_chars=["?", "!"],
keep_punct=True,
)
doc_splitter = DocumentSplitter(
segment_label="sentence",
attr_labels=[], # workaround for issue 199
)
for doc in docs:
sentence_segs = sentence_tokenizer.run([doc.raw_segment])
for sentence_seg in sentence_segs:
doc.anns.add(sentence_seg)
sentence_docs = doc_splitter.run(docs)
return sentence_docs
# Do this for EMEA docs in each split
train_docs_emea_sentences = split_emea_docs(train_docs_emea)
dev_docs_emea_sentences = split_emea_docs(dev_docs_emea)
test_docs_emea_sentences = split_emea_docs(test_docs_emea)
Let’s save this preprocessed version of our dataset in medkit json files, so we can reuse easily another time if needed:
import shutil
from medkit.io import medkit_json
train_docs = train_docs_medline + train_docs_emea_sentences
dev_docs = dev_docs_medline + dev_docs_emea_sentences
test_docs = test_docs_medline + test_docs_emea_sentences
QUAERO_PREPROCESSED_DIR = "QUAERO_preprocessed/"
if os.path.exists(QUAERO_PREPROCESSED_DIR):
shutil.rmtree(QUAERO_PREPROCESSED_DIR)
os.mkdir(QUAERO_PREPROCESSED_DIR)
medkit_json.save_text_documents(train_docs, QUAERO_PREPROCESSED_DIR + "train.jsonl")
medkit_json.save_text_documents(dev_docs, QUAERO_PREPROCESSED_DIR + "dev.jsonl")
medkit_json.save_text_documents(test_docs, QUAERO_PREPROCESSED_DIR + "test.jsonl")
Compute baseline metrics with UMLSMatcher#
Since we have an annotated dataset with entities having UMLS semgroups as
values, we can use it to evaluate the performance of the medkit
UMLSMatcher.
For this we will use the SeqEval library
, which is wrapped in medkit in the
SeqEvalEvaluator class.
It allows us to get the following metrics:
accuracy, the proportion of tokens that were properly labelled (included for the parts of the text which have no entity)
precision, the proportion of correctly identified entities across all predicted entities
recall, the proportion of correctly identified entities across all groundtruth entities
f1 score, which combines both recall and precision
We will use the IOB2 tagging scheme to classify the tokens before computing the metrics. The metrics are computed in strict mode, which means that each token of the entity has to be properly labelled for the entity to be considered as properly identified.
from pprint import pprint
from medkit.text.ner import UMLSMatcher
from medkit.text.metrics.ner import SeqEvalEvaluator
# UMLS semantic groups to take into account
# (QUAERO is only annotated with these)
umls_semgroups = [
"ANAT", # anatomy
"CHEM", # chemical
"DEVI", # device
"DISO", # disorder
"GEOG", # geographic
"LIVB", # living being
"OBJC", # object
"PHEN", # concept
"PHYS", # physiological
"PROC", # procedure
]
umls_matcher = UMLSMatcher(
# Directory containing the UMLS files with all the terms and concepts
umls_dir="../data/umls/2021AB/META/",
# Language to use
language="FRE",
# Where to store the internal terms database of the matcher
cache_dir=".umls_cache/",
# Semantic groups to consider
semgroups=umls_semgroups,
# Don't be case-sensitive
lowercase=True,
# Convert special chars to ASCII before matching
# (same a unicode_sensitive=False for regexp rules)
normalize_unicode=True,
)
# Run the umls matcher on each doc from the test split,
# and keep the correspond predicted entities
# predicted_entities will be a list of list, with one inner list per document
predicted_entities = []
for test_doc in test_docs:
entities = umls_matcher.run([test_doc.raw_segment])
predicted_entities.append(entities)
# Create an evaluator object based on SeqEval
ner_metrics_evaluator = SeqEvalEvaluator(
tagging_scheme="iob2",
return_metrics_by_label=False,
# QUAERO is labelled with 4-letter UMLS semgroups code
# but the UMLSMatcher gives human-readable labels
# so we needed to remap them before computing the metric
labels_remapping={
"anatomy": "ANAT",
"chemical": "CHEM",
"device": "DEVI",
"disorder": "DISO",
"geographic_area": "GEOG",
"living_being": "LIVB",
"object": "OBJC",
"phenomenon": "PHEN",
"physiology": "PHYS",
"procedure": "PROC",
}
)
# Pass it the test documents (which contain the refrence entities)
# along with the predicted entities, and get the metrics in return
umls_matcher_scores = ner_metrics_evaluator.compute(test_docs, predicted_entities)
pprint(umls_matcher_scores)
{
'accuracy': 0.7683723661992512,
'macro_f1-score': 0.4246273234375716,
'macro_precision': 0.5621040123228653,
'macro_recall': 0.3528176174287537,
'support': 4085,
}
We reach a f1-score of approximately 42%.
Finetuning a BERT model for entity matching#
In this example, we show how to fine-tune DrBERT: A PreTrained model in French for Biomedical and Clinical domains to detect entities label with UMLS semantic groups using the medkit Trainer.
DrBert[1] is a French RoBERTa trained in open source corpus of french medical documents for masked language modeling. As mentioned before, we can change the specific task, for example, to classify entities.
Medkit provides simple training tools that make it possible to train or fine-tune some of its operations, such as HFEntityMatcher, through is trainable counterpart HFEntityMatcherTrainable. To use it, we must provide it with a model-based operation to train, a dataset (with train and dev sets), and an optional helper object to compute custom metrics.
Let’s define a trainable instance for this example:
import torch
from medkit.text.ner.hf_entity_matcher import HFEntityMatcher
# Device on which to load the model and peform the training
# -1 for CPU, 0 for the first GPU of the computer, 1 for the 2d GPU, etc
DEVICE = 0 if torch.cuda.is_available() else -1
# Get a trainable component for HFEntityMatcher
trainable_matcher = HFEntityMatcher.make_trainable(
# The DrBERT model on the hugging face hub that we want to fine-une
# cf https://huggingface.co/Dr-BERT/DrBERT-4GB-CP-PubMedBERT
model_name_or_path="Dr-BERT/DrBERT-4GB-CP-PubMedBERT",
# The labels of the entities that will be recognized
labels=umls_semgroups,
# The tagging scheme to use
tagging_scheme="iob2",
# Maximum number of tokens that the model can receive at once
tokenizer_max_length=512,
# The device on which to train the model
device=DEVICE,
)
At this point, we have prepared the data and the component to fine-tune. All we need to do is define the trainer with its configuration
from medkit.training import TrainerConfig, Trainer
from medkit.text.metrics.ner import SeqEvalMetricsComputer
CHECKPOINT_DIR = "checkpoints/"
# Prepare the config for the trainer class
trainer_config = TrainerConfig(
# Where to save the model weights after training
output_dir=CHECKPOINT_DIR,
# Learning rate to use
learning_rate=2e-6,
# Number of training epochs
nb_training_epochs=1,
# Number of documents (ie sentences) per batch
batch_size=5,
)
# Prepare the object that will compute the NER metrics at the end of each epoch
# (this is a variant of the SeqEvalMetricsEvaluator that we used earlier, that
# can be used during training)
ner_metrics_computer = SeqEvalMetricsComputer(
id_to_label=trainable_matcher.id_to_label,
tagging_scheme=trainable_matcher.tagging_scheme,
return_metrics_by_label=False,
)
# Init a trainer object giving it:
trainer = Trainer(
# its configuration
config=trainer_config,
# the component (ie model) it has to train
component=trainable_matcher,
# the training and dev sets
train_data=train_docs,
eval_data=dev_docs,
# the helper object that computes the metrics at each epoch
metrics_computer=ner_metrics_computer,
)
We can now run the training loop with trainer.train(). It returns a dictionary
with the training history and saves a checkpoint with the tuned model:
# Run training and keep history of losses and metrics
history = trainer.train()
Let’s take a look at how the metrics evolved during the training:
recall = [epoch["eval"]["macro_recall"] for epoch in history]
precision = [epoch["eval"]["macro_precision"] for epoch in history]
f1_score = [epoch["eval"]["macro_f1-score"] for epoch in history]
plt.plot(recall, label="recall")
plt.plot(precision, label="precision")
plt.plot(f1_score, label="f1_score")
plt.legend()
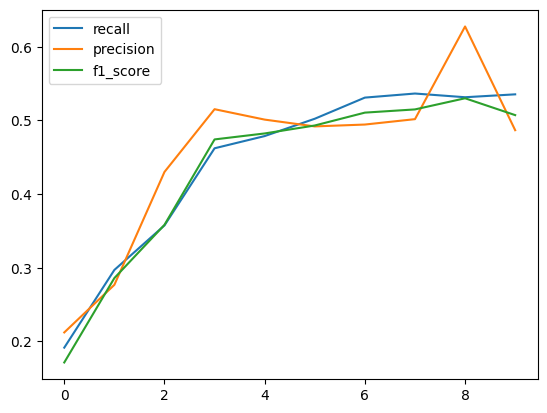
After 10 epochs, we should reach a f1-score around 50% on the dev split. Let’s
look at the final metrics on the test split. For this we will reinstantiate an
HFEntityMatcher with the last checkpoint:
from glob import glob
# Retrieve best checkpoint and use it to instantiate the HuggingFace entity matcher
checkpoint_path = sorted(glob(CHECKPOINT_DIR + "/checkpoint_*"))[-1]
trained_drbert_matcher = HFEntityMatcher(model=checkpoint_path, device=DEVICE)
# Get the predicted entities for each test document
predicted_entities = []
for test_doc in test_docs:
entities = trained_drbert_matcher.run([test_doc.raw_segment])
predicted_entities.append(entities)
# Compute NER metrics on test docs
metrics = ner_metrics_evaluator.compute(test_docs, predicted_entities)
pprint(metrics)
{
'accuracy': 0.8604727993539387,
'macro_f1-score': 0.48133173293574166,
'macro_precision': 0.5292726724036432,
'macro_recall': 0.4690750888795575,
'support': 4085,
}
Our fine-tuned BERT model has a better f1-score than the fuzzy simstring matcher (48% vs 42%), thanks to its better recall (47% vs 35%).
