Comparative evaluation of drug recognition methods#
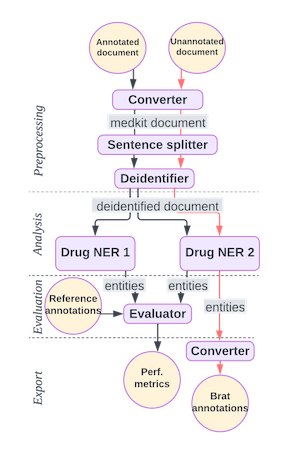
This pipeline compares two tools (NER1 and NER2) for recognizing drug names in clinical texts, compares their performance, and outputs two texts annotated with the tool evaluated as the best performer.
Overview of the pipeline#
Data preparation#
Download two clinical texts, with drug entites manually annotated.
import os
import tarfile
import tempfile
from pathlib import Path
# path to local data
extract_to = Path(tempfile.mkdtemp())
data_tarfile = Path.cwd() / "data.tar.gz"
# download and extract
tarfile.open(name=data_tarfile, mode="r|gz").extractall(extract_to)
data_dir = extract_to / "data"
print(f"Data dir: {data_dir}")
Data dir: /tmp/tmpyoac7euf/data
Read text documents with medkit
from medkit.core.text import TextDocument
doc_dir = data_dir / "mtsamplesen" / "annotated_doc"
docs = TextDocument.from_dir(path=doc_dir, pattern='[A-Z0-9].txt', encoding='utf-8')
print(docs[0].text)
DISCHARGE DIAGNOSES:
1. Gram-negative rod bacteremia, final identification and susceptibilities still pending.
2. History of congenital genitourinary abnormalities with multiple surgeries before the 5th grade.
3. History of urinary tract infections of pyelonephritis.
OPERATIONS PERFORMED: Chest x-ray July 24, 2007, that was normal. Transesophageal echocardiogram July 27, 2007, that was normal. No evidence of vegetations. CT scan of the abdomen and pelvis July 27, 2007, that revealed multiple small cysts in the liver, the largest measuring 9 mm. There were 2-3 additional tiny cysts in the right lobe. The remainder of the CT scan was normal.
HISTORY OF PRESENT ILLNESS: Briefly, the patient is a 26-year-old white female with a history of fevers. For further details of the admission, please see the previously dictated history and physical.
HOSPITAL COURSE: Gram-negative rod bacteremia. The patient was admitted to the hospital with suspicion of endocarditis given the fact that she had fever, septicemia, and Osler nodes on her fingers. The patient had a transthoracic echocardiogram as an outpatient, which was equivocal, but a transesophageal echocardiogram here in the hospital was normal with no evidence of vegetations. The microbiology laboratory stated that the Gram-negative rod appeared to be anaerobic, thus raising the possibility of organisms like bacteroides. The patient does have a history of congenital genitourinary abnormalities which were surgically corrected before the fifth grade. We did a CT scan of the abdomen and pelvis, which only showed some benign appearing cysts in the liver. There was nothing remarkable as far as her kidneys, ureters, or bladder were concerned. I spoke with Dr. Leclerc of infectious diseases, and Dr. Leclerc asked me to talk to the patient about any contact with animals, given the fact that we have had a recent outbreak of tularemia here in Utah. Much to my surprise, the patient told me that she had multiple pet rats at home, which she was constantly in contact with. I ordered tularemia and leptospirosis serologies on the advice of Dr. Leclerc, and as of the day after discharge, the results of the microbiology still are not back yet. The patient, however, appeared to be responding well to levofloxacin. I gave her a 2-week course of 750 mg a day of levofloxacin, and I have instructed her to follow up with Dr. Leclerc in the meantime. Hopefully by then we will have a final identification and susceptibility on the organism and the tularemia and leptospirosis serologies will return. A thought of ours was to add doxycycline, but again the patient clinically appeared to be responding to the levofloxacin. In addition, I told the patient that it would be my recommendation to get rid of the rats. I told her that if indeed the rats were carriers of infection and she received a zoonotic infection from exposure to the rats, that she could be in ongoing continuing danger and her children could also potentially be exposed to a potentially lethal infection. I told her very clearly that she should, indeed, get rid of the animals. The patient seemed reluctant to do so at first, but I believe with some coercion from her family, that she finally came to the realization that this was a recommendation worth following.
DISPOSITION
DISCHARGE INSTRUCTIONS: Activity is as tolerated. Diet is as tolerated.
MEDICATIONS: Levaquin 750 mg daily x14 days.
Followup is with Dr. Leclerc of infectious diseases. I gave the patient the phone number to call on Monday for an appointment. Additional followup is also with Dr. Leclerc, her primary care physician. Please note that 40 minutes was spent in the discharge.
Pipeline definition#
Create and run a three-step doc pipeline that:
Split sentences in texts
Run PII detection for deidentification
Recognize drug entities with NER1: a dictionnary-based approach named UMLSMatcher
Recognize drug entities with NER2: a Transformer-based approach, see https://huggingface.co/samrawal/bert-large-uncased_med-ner
Sentence tokenizer#
from medkit.text.segmentation import SentenceTokenizer
# By default, SentenceTokenizer will use a list of punctuation chars to detect sentences.
sentence_tokenizer = SentenceTokenizer(
# Label of the segments created and returned by the operation
output_label="sentence",
# Keep the punctuation character inside the sentence segments
keep_punct=True,
# Also split on newline chars, not just punctuation characters
split_on_newlines=True,
)
PII detector#
from medkit.text.deid import PIIDetector
pii_detector = PIIDetector(name="deid")
Collecting en-core-web-lg==3.7.1
Downloading https://github.com/explosion/spacy-models/releases/download/en_core_web_lg-3.7.1/en_core_web_lg-3.7.1-py3-none-any.whl (587.7 MB)
?25l ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0.0/587.7 MB ? eta -:--:--
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 3.3/587.7 MB 99.2 MB/s eta 0:00:06
╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 9.0/587.7 MB 132.2 MB/s eta 0:00:05
╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 15.3/587.7 MB 178.1 MB/s eta 0:00:04
━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 21.5/587.7 MB 182.2 MB/s eta 0:00:04
━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 27.9/587.7 MB 184.6 MB/s eta 0:00:04
━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 34.2/587.7 MB 184.4 MB/s eta 0:00:04
━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 40.4/587.7 MB 182.9 MB/s eta 0:00:03
━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 46.4/587.7 MB 176.8 MB/s eta 0:00:04
━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 52.6/587.7 MB 179.3 MB/s eta 0:00:03
━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 58.9/587.7 MB 181.2 MB/s eta 0:00:03
━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 65.2/587.7 MB 183.1 MB/s eta 0:00:03
━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 71.3/587.7 MB 181.1 MB/s eta 0:00:03
━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 77.5/587.7 MB 179.2 MB/s eta 0:00:03
━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 83.7/587.7 MB 179.2 MB/s eta 0:00:03
━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 89.9/587.7 MB 179.5 MB/s eta 0:00:03
━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 96.2/587.7 MB 181.7 MB/s eta 0:00:03
━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 102.4/587.7 MB 182.1 MB/s eta 0:00:03
━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 108.4/587.7 MB 178.5 MB/s eta 0:00:03
━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 114.6/587.7 MB 178.9 MB/s eta 0:00:03
━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 120.8/587.7 MB 180.1 MB/s eta 0:00:03
━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 127.0/587.7 MB 178.3 MB/s eta 0:00:03
━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 133.3/587.7 MB 180.0 MB/s eta 0:00:03
━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 139.4/587.7 MB 181.4 MB/s eta 0:00:03
━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━━ 145.8/587.7 MB 182.3 MB/s eta 0:00:03
━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━ 151.9/587.7 MB 181.9 MB/s eta 0:00:03
━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━━ 158.1/587.7 MB 178.8 MB/s eta 0:00:03
━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━━ 164.4/587.7 MB 180.2 MB/s eta 0:00:03
━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━━ 170.6/587.7 MB 183.9 MB/s eta 0:00:03
━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━━ 177.0/587.7 MB 184.1 MB/s eta 0:00:03
━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━ 183.4/587.7 MB 186.5 MB/s eta 0:00:03
━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━━ 189.4/587.7 MB 180.9 MB/s eta 0:00:03
━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━━ 195.5/587.7 MB 178.5 MB/s eta 0:00:03
━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━━ 201.7/587.7 MB 177.7 MB/s eta 0:00:03
━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━ 208.0/587.7 MB 181.6 MB/s eta 0:00:03
━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━━ 214.4/587.7 MB 183.7 MB/s eta 0:00:03
━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━━ 220.7/587.7 MB 184.0 MB/s eta 0:00:02
━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━━ 226.9/587.7 MB 181.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━━ 233.3/587.7 MB 184.4 MB/s eta 0:00:02
━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━━ 239.7/587.7 MB 185.7 MB/s eta 0:00:02
━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━ 246.5/587.7 MB 194.6 MB/s eta 0:00:02
━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━━ 253.1/587.7 MB 195.4 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━━ 259.9/587.7 MB 193.5 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━━ 267.0/587.7 MB 201.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━━ 273.8/587.7 MB 204.6 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━━ 280.5/587.7 MB 194.4 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━ 286.9/587.7 MB 189.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━━ 293.1/587.7 MB 182.6 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━━ 299.3/587.7 MB 181.6 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━━ 305.8/587.7 MB 186.0 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━━ 312.0/587.7 MB 185.4 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━ 318.2/587.7 MB 178.8 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━━ 324.5/587.7 MB 181.4 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━━ 330.7/587.7 MB 182.8 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━━ 336.8/587.7 MB 179.3 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━ 343.0/587.7 MB 178.7 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━━ 349.2/587.7 MB 180.5 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━━ 355.5/587.7 MB 181.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━━ 361.7/587.7 MB 181.9 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━━ 368.0/587.7 MB 182.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━ 374.2/587.7 MB 182.7 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━━ 380.5/587.7 MB 182.8 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━━ 386.9/587.7 MB 183.8 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━━ 392.9/587.7 MB 178.0 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━━ 399.1/587.7 MB 180.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━ 405.4/587.7 MB 182.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━━ 411.6/587.7 MB 182.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━━ 417.9/587.7 MB 180.1 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━━ 423.9/587.7 MB 179.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━ 430.0/587.7 MB 176.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━━ 436.1/587.7 MB 177.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━━ 442.3/587.7 MB 178.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━━ 448.4/587.7 MB 178.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━ 454.5/587.7 MB 178.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━━ 460.5/587.7 MB 175.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━━ 466.7/587.7 MB 176.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━━ 472.8/587.7 MB 178.1 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━━ 479.2/587.7 MB 183.1 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━ 485.3/587.7 MB 180.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━ 489.3/587.7 MB 154.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━ 490.9/587.7 MB 107.4 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━━ 494.1/587.7 MB 89.1 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━━ 497.6/587.7 MB 80.2 MB/s eta 0:00:02
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━ 502.4/587.7 MB 110.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━ 503.8/587.7 MB 90.5 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━ 507.2/587.7 MB 91.6 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━━ 509.3/587.7 MB 80.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━ 512.9/587.7 MB 87.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━━ 517.5/587.7 MB 99.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━ 519.5/587.7 MB 96.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━ 522.2/587.7 MB 83.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━━ 524.6/587.7 MB 84.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━ 527.2/587.7 MB 71.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━ 530.1/587.7 MB 76.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━━ 533.1/587.7 MB 85.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━ 535.8/587.7 MB 82.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━━ 538.5/587.7 MB 79.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━ 542.2/587.7 MB 84.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━━ 545.3/587.7 MB 88.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━ 549.3/587.7 MB 106.9 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━━ 552.8/587.7 MB 104.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━ 556.0/587.7 MB 105.8 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━ 559.1/587.7 MB 93.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺━ 562.4/587.7 MB 93.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━ 565.7/587.7 MB 94.2 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸━ 568.5/587.7 MB 87.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺ 572.8/587.7 MB 99.7 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╺ 576.9/587.7 MB 111.3 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 580.9/587.7 MB 113.1 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 585.0/587.7 MB 115.9 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 587.7/587.7 MB 127.0 MB/s eta 0:00:01
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 587.7/587.7 MB 4.3 MB/s eta 0:00:00
?25h
Requirement already satisfied: spacy<3.8.0,>=3.7.2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from en-core-web-lg==3.7.1) (3.7.6)
Requirement already satisfied: spacy-legacy<3.1.0,>=3.0.11 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.0.12)
Requirement already satisfied: spacy-loggers<2.0.0,>=1.0.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.0.5)
Requirement already satisfied: murmurhash<1.1.0,>=0.28.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.0.10)
Requirement already satisfied: cymem<2.1.0,>=2.0.2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.0.8)
Requirement already satisfied: preshed<3.1.0,>=3.0.2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.0.9)
Requirement already satisfied: thinc<8.3.0,>=8.2.2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (8.2.5)
Requirement already satisfied: wasabi<1.2.0,>=0.9.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.1.3)
Requirement already satisfied: srsly<3.0.0,>=2.4.3 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.4.8)
Requirement already satisfied: catalogue<2.1.0,>=2.0.6 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.0.10)
Requirement already satisfied: weasel<0.5.0,>=0.1.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.4.1)
Requirement already satisfied: typer<1.0.0,>=0.3.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.12.5)
Requirement already satisfied: tqdm<5.0.0,>=4.38.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (4.66.5)
Requirement already satisfied: requests<3.0.0,>=2.13.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.32.3)
Requirement already satisfied: pydantic!=1.8,!=1.8.1,<3.0.0,>=1.7.4 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.9.2)
Requirement already satisfied: jinja2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.1.4)
Requirement already satisfied: setuptools in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (69.5.1)
Requirement already satisfied: packaging>=20.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (24.1)
Requirement already satisfied: langcodes<4.0.0,>=3.2.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.4.1)
Requirement already satisfied: numpy>=1.19.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.26.4)
Requirement already satisfied: language-data>=1.2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from langcodes<4.0.0,>=3.2.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.2.0)
Requirement already satisfied: annotated-types>=0.6.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from pydantic!=1.8,!=1.8.1,<3.0.0,>=1.7.4->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.7.0)
Requirement already satisfied: pydantic-core==2.23.4 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from pydantic!=1.8,!=1.8.1,<3.0.0,>=1.7.4->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.23.4)
Requirement already satisfied: typing-extensions>=4.6.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from pydantic!=1.8,!=1.8.1,<3.0.0,>=1.7.4->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (4.12.2)
Requirement already satisfied: charset-normalizer<4,>=2 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.13.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.3.2)
Requirement already satisfied: idna<4,>=2.5 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.13.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.10)
Requirement already satisfied: urllib3<3,>=1.21.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.13.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.2.3)
Requirement already satisfied: certifi>=2017.4.17 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from requests<3.0.0,>=2.13.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2024.8.30)
Requirement already satisfied: blis<0.8.0,>=0.7.8 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from thinc<8.3.0,>=8.2.2->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.7.11)
Requirement already satisfied: confection<1.0.0,>=0.0.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from thinc<8.3.0,>=8.2.2->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.1.5)
Requirement already satisfied: click>=8.0.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (8.1.7)
Requirement already satisfied: shellingham>=1.3.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.5.4)
Requirement already satisfied: rich>=10.11.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (13.8.1)
Requirement already satisfied: cloudpathlib<1.0.0,>=0.7.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from weasel<0.5.0,>=0.1.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.19.0)
Requirement already satisfied: smart-open<8.0.0,>=5.2.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from weasel<0.5.0,>=0.1.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (7.0.4)
Requirement already satisfied: MarkupSafe>=2.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from jinja2->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.1.5)
Requirement already satisfied: marisa-trie>=0.7.7 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from language-data>=1.2->langcodes<4.0.0,>=3.2.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.2.0)
Requirement already satisfied: markdown-it-py>=2.2.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from rich>=10.11.0->typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (3.0.0)
Requirement already satisfied: pygments<3.0.0,>=2.13.0 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from rich>=10.11.0->typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (2.18.0)
Requirement already satisfied: wrapt in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from smart-open<8.0.0,>=5.2.1->weasel<0.5.0,>=0.1.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (1.16.0)
Requirement already satisfied: mdurl~=0.1 in /home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages (from markdown-it-py>=2.2.0->rich>=10.11.0->typer<1.0.0,>=0.3.0->spacy<3.8.0,>=3.7.2->en-core-web-lg==3.7.1) (0.1.2)
Installing collected packages: en-core-web-lg
Successfully installed en-core-web-lg-3.7.1
[notice] A new release of pip is available: 24.0 -> 24.2
[notice] To update, run: pip install --upgrade pip
✔ Download and installation successful
You can now load the package via spacy.load('en_core_web_lg')
⚠ Restart to reload dependencies
If you are in a Jupyter or Colab notebook, you may need to restart Python in
order to load all the package's dependencies. You can do this by selecting the
'Restart kernel' or 'Restart runtime' option.
Dictionnary-based drug recognizer#
import shutil
from medkit.text.ner import UMLSMatcher
umls_data_dir = data_dir / "UMLS" / "2023AB" / "META"
umls_cache_dir = Path.cwd() / ".umls_cache"
shutil.rmtree(umls_cache_dir, ignore_errors=True)
umls_matcher = UMLSMatcher(
# Directory containing the UMLS files with terms and concepts
umls_dir=umls_data_dir,
# Language to use (English)
language="ENG",
# Where to store the temp term database of the matcher
cache_dir=umls_cache_dir,
# Semantic groups to consider
semgroups=["CHEM"],
# Don't be case-sensitive
lowercase=True,
# Convert special chars to ASCII before matching
normalize_unicode=True,
name="NER1"
)
/home/runner/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/tqdm/auto.py:21: TqdmWarning: IProgress not found. Please update jupyter and ipywidgets. See https://ipywidgets.readthedocs.io/en/stable/user_install.html
from .autonotebook import tqdm as notebook_tqdm
0%| | 0.00/13.9k [00:00<?, ?B/s]
100%|██████████| 13.9k/13.9k [00:00<00:00, 1.44MB/s]
Transformer-based drug recognizer#
from medkit.text.ner.hf_entity_matcher import HFEntityMatcher
# an alternate model: "Clinical-AI-Apollo/Medical-NER"
bert_matcher = HFEntityMatcher(
model="samrawal/bert-large-uncased_med-ner", name="NER2"
)
---------------------------------------------------------------------------
KeyboardInterrupt Traceback (most recent call last)
Cell In[6], line 4
1 from medkit.text.ner.hf_entity_matcher import HFEntityMatcher
3 # an alternate model: "Clinical-AI-Apollo/Medical-NER"
----> 4 bert_matcher = HFEntityMatcher(
5 model="samrawal/bert-large-uncased_med-ner", name="NER2"
6 )
File ~/work/medkit/medkit/medkit/text/ner/hf_entity_matcher.py:91, in HFEntityMatcher.__init__(self, model, aggregation_strategy, attrs_to_copy, device, batch_size, hf_auth_token, cache_dir, name, uid)
84 msg = (
85 f"Model {self.model} is not associated to a"
86 " token-classification/ner task and cannot be used with"
87 " HFEntityMatcher"
88 )
89 raise ValueError(msg)
---> 91 self._pipeline = transformers.pipeline(
92 task="token-classification",
93 model=self.model,
94 aggregation_strategy=aggregation_strategy,
95 pipeline_class=transformers.TokenClassificationPipeline,
96 device=device,
97 batch_size=batch_size,
98 token=hf_auth_token,
99 model_kwargs={"cache_dir": cache_dir},
100 )
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/transformers/pipelines/__init__.py:994, in pipeline(task, model, config, tokenizer, feature_extractor, image_processor, framework, revision, use_fast, token, device, device_map, torch_dtype, trust_remote_code, model_kwargs, pipeline_class, **kwargs)
991 tokenizer_kwargs = model_kwargs.copy()
992 tokenizer_kwargs.pop("torch_dtype", None)
--> 994 tokenizer = AutoTokenizer.from_pretrained(
995 tokenizer_identifier, use_fast=use_fast, _from_pipeline=task, **hub_kwargs, **tokenizer_kwargs
996 )
998 if load_image_processor:
999 # Try to infer image processor from model or config name (if provided as str)
1000 if image_processor is None:
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/transformers/models/auto/tokenization_auto.py:916, in AutoTokenizer.from_pretrained(cls, pretrained_model_name_or_path, *inputs, **kwargs)
913 tokenizer_class_py, tokenizer_class_fast = TOKENIZER_MAPPING[type(config)]
915 if tokenizer_class_fast and (use_fast or tokenizer_class_py is None):
--> 916 return tokenizer_class_fast.from_pretrained(pretrained_model_name_or_path, *inputs, **kwargs)
917 else:
918 if tokenizer_class_py is not None:
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/transformers/tokenization_utils_base.py:2227, in PreTrainedTokenizerBase.from_pretrained(cls, pretrained_model_name_or_path, cache_dir, force_download, local_files_only, token, revision, trust_remote_code, *init_inputs, **kwargs)
2225 resolved_vocab_files[file_id] = download_url(file_path, proxies=proxies)
2226 else:
-> 2227 resolved_vocab_files[file_id] = cached_file(
2228 pretrained_model_name_or_path,
2229 file_path,
2230 cache_dir=cache_dir,
2231 force_download=force_download,
2232 proxies=proxies,
2233 resume_download=resume_download,
2234 local_files_only=local_files_only,
2235 token=token,
2236 user_agent=user_agent,
2237 revision=revision,
2238 subfolder=subfolder,
2239 _raise_exceptions_for_gated_repo=False,
2240 _raise_exceptions_for_missing_entries=False,
2241 _raise_exceptions_for_connection_errors=False,
2242 _commit_hash=commit_hash,
2243 )
2244 commit_hash = extract_commit_hash(resolved_vocab_files[file_id], commit_hash)
2246 if len(unresolved_files) > 0:
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/transformers/utils/hub.py:402, in cached_file(path_or_repo_id, filename, cache_dir, force_download, resume_download, proxies, token, revision, local_files_only, subfolder, repo_type, user_agent, _raise_exceptions_for_gated_repo, _raise_exceptions_for_missing_entries, _raise_exceptions_for_connection_errors, _commit_hash, **deprecated_kwargs)
399 user_agent = http_user_agent(user_agent)
400 try:
401 # Load from URL or cache if already cached
--> 402 resolved_file = hf_hub_download(
403 path_or_repo_id,
404 filename,
405 subfolder=None if len(subfolder) == 0 else subfolder,
406 repo_type=repo_type,
407 revision=revision,
408 cache_dir=cache_dir,
409 user_agent=user_agent,
410 force_download=force_download,
411 proxies=proxies,
412 resume_download=resume_download,
413 token=token,
414 local_files_only=local_files_only,
415 )
416 except GatedRepoError as e:
417 resolved_file = _get_cache_file_to_return(path_or_repo_id, full_filename, cache_dir, revision)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/utils/_deprecation.py:101, in _deprecate_arguments.<locals>._inner_deprecate_positional_args.<locals>.inner_f(*args, **kwargs)
99 message += "\n\n" + custom_message
100 warnings.warn(message, FutureWarning)
--> 101 return f(*args, **kwargs)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/utils/_validators.py:114, in validate_hf_hub_args.<locals>._inner_fn(*args, **kwargs)
111 if check_use_auth_token:
112 kwargs = smoothly_deprecate_use_auth_token(fn_name=fn.__name__, has_token=has_token, kwargs=kwargs)
--> 114 return fn(*args, **kwargs)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:1232, in hf_hub_download(repo_id, filename, subfolder, repo_type, revision, library_name, library_version, cache_dir, local_dir, user_agent, force_download, proxies, etag_timeout, token, local_files_only, headers, endpoint, legacy_cache_layout, resume_download, force_filename, local_dir_use_symlinks)
1212 return _hf_hub_download_to_local_dir(
1213 # Destination
1214 local_dir=local_dir,
(...)
1229 local_files_only=local_files_only,
1230 )
1231 else:
-> 1232 return _hf_hub_download_to_cache_dir(
1233 # Destination
1234 cache_dir=cache_dir,
1235 # File info
1236 repo_id=repo_id,
1237 filename=filename,
1238 repo_type=repo_type,
1239 revision=revision,
1240 # HTTP info
1241 endpoint=endpoint,
1242 etag_timeout=etag_timeout,
1243 headers=headers,
1244 proxies=proxies,
1245 token=token,
1246 # Additional options
1247 local_files_only=local_files_only,
1248 force_download=force_download,
1249 )
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:1295, in _hf_hub_download_to_cache_dir(cache_dir, repo_id, filename, repo_type, revision, endpoint, etag_timeout, headers, proxies, token, local_files_only, force_download)
1291 return pointer_path
1293 # Try to get metadata (etag, commit_hash, url, size) from the server.
1294 # If we can't, a HEAD request error is returned.
-> 1295 (url_to_download, etag, commit_hash, expected_size, head_call_error) = _get_metadata_or_catch_error(
1296 repo_id=repo_id,
1297 filename=filename,
1298 repo_type=repo_type,
1299 revision=revision,
1300 endpoint=endpoint,
1301 proxies=proxies,
1302 etag_timeout=etag_timeout,
1303 headers=headers,
1304 token=token,
1305 local_files_only=local_files_only,
1306 storage_folder=storage_folder,
1307 relative_filename=relative_filename,
1308 )
1310 # etag can be None for several reasons:
1311 # 1. we passed local_files_only.
1312 # 2. we don't have a connection
(...)
1318 # If the specified revision is a commit hash, look inside "snapshots".
1319 # If the specified revision is a branch or tag, look inside "refs".
1320 if head_call_error is not None:
1321 # Couldn't make a HEAD call => let's try to find a local file
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:1746, in _get_metadata_or_catch_error(repo_id, filename, repo_type, revision, endpoint, proxies, etag_timeout, headers, token, local_files_only, relative_filename, storage_folder)
1744 try:
1745 try:
-> 1746 metadata = get_hf_file_metadata(
1747 url=url, proxies=proxies, timeout=etag_timeout, headers=headers, token=token
1748 )
1749 except EntryNotFoundError as http_error:
1750 if storage_folder is not None and relative_filename is not None:
1751 # Cache the non-existence of the file
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/utils/_validators.py:114, in validate_hf_hub_args.<locals>._inner_fn(*args, **kwargs)
111 if check_use_auth_token:
112 kwargs = smoothly_deprecate_use_auth_token(fn_name=fn.__name__, has_token=has_token, kwargs=kwargs)
--> 114 return fn(*args, **kwargs)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:1666, in get_hf_file_metadata(url, token, proxies, timeout, library_name, library_version, user_agent, headers)
1663 headers["Accept-Encoding"] = "identity" # prevent any compression => we want to know the real size of the file
1665 # Retrieve metadata
-> 1666 r = _request_wrapper(
1667 method="HEAD",
1668 url=url,
1669 headers=headers,
1670 allow_redirects=False,
1671 follow_relative_redirects=True,
1672 proxies=proxies,
1673 timeout=timeout,
1674 )
1675 hf_raise_for_status(r)
1677 # Return
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:364, in _request_wrapper(method, url, follow_relative_redirects, **params)
362 # Recursively follow relative redirects
363 if follow_relative_redirects:
--> 364 response = _request_wrapper(
365 method=method,
366 url=url,
367 follow_relative_redirects=False,
368 **params,
369 )
371 # If redirection, we redirect only relative paths.
372 # This is useful in case of a renamed repository.
373 if 300 <= response.status_code <= 399:
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/file_download.py:387, in _request_wrapper(method, url, follow_relative_redirects, **params)
384 return response
386 # Perform request and return if status_code is not in the retry list.
--> 387 response = get_session().request(method=method, url=url, **params)
388 hf_raise_for_status(response)
389 return response
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/requests/sessions.py:589, in Session.request(self, method, url, params, data, headers, cookies, files, auth, timeout, allow_redirects, proxies, hooks, stream, verify, cert, json)
584 send_kwargs = {
585 "timeout": timeout,
586 "allow_redirects": allow_redirects,
587 }
588 send_kwargs.update(settings)
--> 589 resp = self.send(prep, **send_kwargs)
591 return resp
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/requests/sessions.py:703, in Session.send(self, request, **kwargs)
700 start = preferred_clock()
702 # Send the request
--> 703 r = adapter.send(request, **kwargs)
705 # Total elapsed time of the request (approximately)
706 elapsed = preferred_clock() - start
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/huggingface_hub/utils/_http.py:93, in UniqueRequestIdAdapter.send(self, request, *args, **kwargs)
91 """Catch any RequestException to append request id to the error message for debugging."""
92 try:
---> 93 return super().send(request, *args, **kwargs)
94 except requests.RequestException as e:
95 request_id = request.headers.get(X_AMZN_TRACE_ID)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/requests/adapters.py:667, in HTTPAdapter.send(self, request, stream, timeout, verify, cert, proxies)
664 timeout = TimeoutSauce(connect=timeout, read=timeout)
666 try:
--> 667 resp = conn.urlopen(
668 method=request.method,
669 url=url,
670 body=request.body,
671 headers=request.headers,
672 redirect=False,
673 assert_same_host=False,
674 preload_content=False,
675 decode_content=False,
676 retries=self.max_retries,
677 timeout=timeout,
678 chunked=chunked,
679 )
681 except (ProtocolError, OSError) as err:
682 raise ConnectionError(err, request=request)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/urllib3/connectionpool.py:789, in HTTPConnectionPool.urlopen(self, method, url, body, headers, retries, redirect, assert_same_host, timeout, pool_timeout, release_conn, chunked, body_pos, preload_content, decode_content, **response_kw)
786 response_conn = conn if not release_conn else None
788 # Make the request on the HTTPConnection object
--> 789 response = self._make_request(
790 conn,
791 method,
792 url,
793 timeout=timeout_obj,
794 body=body,
795 headers=headers,
796 chunked=chunked,
797 retries=retries,
798 response_conn=response_conn,
799 preload_content=preload_content,
800 decode_content=decode_content,
801 **response_kw,
802 )
804 # Everything went great!
805 clean_exit = True
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/urllib3/connectionpool.py:536, in HTTPConnectionPool._make_request(self, conn, method, url, body, headers, retries, timeout, chunked, response_conn, preload_content, decode_content, enforce_content_length)
534 # Receive the response from the server
535 try:
--> 536 response = conn.getresponse()
537 except (BaseSSLError, OSError) as e:
538 self._raise_timeout(err=e, url=url, timeout_value=read_timeout)
File ~/.local/share/hatch/env/virtual/medkit-lib/KiEdgqfH/docs/lib/python3.11/site-packages/urllib3/connection.py:507, in HTTPConnection.getresponse(self)
504 from .response import HTTPResponse
506 # Get the response from http.client.HTTPConnection
--> 507 httplib_response = super().getresponse()
509 try:
510 assert_header_parsing(httplib_response.msg)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/http/client.py:1395, in HTTPConnection.getresponse(self)
1393 try:
1394 try:
-> 1395 response.begin()
1396 except ConnectionError:
1397 self.close()
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/http/client.py:325, in HTTPResponse.begin(self)
323 # read until we get a non-100 response
324 while True:
--> 325 version, status, reason = self._read_status()
326 if status != CONTINUE:
327 break
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/http/client.py:286, in HTTPResponse._read_status(self)
285 def _read_status(self):
--> 286 line = str(self.fp.readline(_MAXLINE + 1), "iso-8859-1")
287 if len(line) > _MAXLINE:
288 raise LineTooLong("status line")
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/socket.py:718, in SocketIO.readinto(self, b)
716 while True:
717 try:
--> 718 return self._sock.recv_into(b)
719 except timeout:
720 self._timeout_occurred = True
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/ssl.py:1314, in SSLSocket.recv_into(self, buffer, nbytes, flags)
1310 if flags != 0:
1311 raise ValueError(
1312 "non-zero flags not allowed in calls to recv_into() on %s" %
1313 self.__class__)
-> 1314 return self.read(nbytes, buffer)
1315 else:
1316 return super().recv_into(buffer, nbytes, flags)
File /opt/hostedtoolcache/Python/3.11.10/x64/lib/python3.11/ssl.py:1166, in SSLSocket.read(self, len, buffer)
1164 try:
1165 if buffer is not None:
-> 1166 return self._sslobj.read(len, buffer)
1167 else:
1168 return self._sslobj.read(len)
KeyboardInterrupt:
Pipeline assembly#
from medkit.core import DocPipeline, Pipeline, PipelineStep
pipeline = Pipeline(
steps=[
PipelineStep(sentence_tokenizer, input_keys=["full_text"], output_keys=["sentence"]),
PipelineStep(pii_detector, input_keys=["sentence"], output_keys=["sentence_"]),
PipelineStep(umls_matcher, input_keys=["sentence_"], output_keys=["ner1_drug"]),
PipelineStep(bert_matcher, input_keys=["sentence_"], output_keys=["ner2_drug"]),
],
input_keys=["full_text"],
output_keys=["sentence_", "ner1_drug", "ner2_drug"],
)
doc_pipeline = DocPipeline(pipeline=pipeline)
doc_pipeline.run(docs)
Performance evaluation#
from medkit.io.brat import BratInputConverter
# Load text with annotations in medkit (our ground truth)
brat_converter = BratInputConverter()
ref_docs = brat_converter.load(doc_dir)
# Display selected drug annotations
for ann in ref_docs[0].anns.get(label="Drug"):
print(f"{ann.text} in {ann.spans}")
## Compute some stats
print(f"Number of documents: {len(docs)}")
for i, doc in enumerate(docs):
print(f"Document {doc.uid}:")
# On annotations made by NER1 and NER2
sentence_nb = len(doc.anns.get(label="sentence"))
print(f"\t{sentence_nb} sentences,")
ner1_drug_nb = len(doc.anns.get(label="chemical"))
print(f"\t{ner1_drug_nb} drugs found with NER1,")
ner2_drug_nb = len(doc.anns.get(label="m"))
print(f"\t{ner2_drug_nb} drugs found with NER2,")
# On the manual annotation (our ground truth)
gt_nb = len(ref_docs[i].anns.get(label="Drug"))
print(f"\t{gt_nb} drugs manually annotated.")
## Evaluate performance metrics of the NER1 and NER2 tools
from medkit.text.metrics.ner import SeqEvalEvaluator
import pandas as pd
def results_to_df(_results, _title):
results_list = list(_results.items())
arranged_results = {"Entities": ['P', 'R', 'F1']}
accuracy = round(results_list[4][1], 2)
for i in range(5, len(results_list), 4):
key = results_list[i][0][:-10]
arranged_results[key] = [round(results_list[n][1], 2) for n in [i, i + 1, i + 2]]
df = pd.DataFrame(arranged_results, index=[f"{_title} (acc={accuracy})", '', '']).T
return df
predicted_entities1=[]
predicted_entities2=[]
dfs = []
for doc in docs:
predicted_entities1.append(doc.anns.get(label="chemical"))
predicted_entities2.append(doc.anns.get(label="m"))
# Annotations of NER1 are labelled as 'chemical', NER2 as 'm', but as 'Drug' in the ground truth
# The following dic enables remappings various labels of the same type of entites
remapping= {"chemical": "Drug", "m": "Drug"}
evaluator = SeqEvalEvaluator(return_metrics_by_label=True, average='weighted', labels_remapping=remapping)
# eval of NER2
results1 = evaluator.compute(ref_docs, predicted_entities1)
dfs.append(results_to_df(_results=results1, _title="NER1"))
#print(results_to_df(_results=results1, _title="umls_matcher"))
# eval of NER2
results2 = evaluator.compute(ref_docs, predicted_entities2)
dfs.append(results_to_df(_results=results2, _title="NER2"))
print(pd.concat(dfs, axis=1))
## Read new unannotated documents
## Write annotations of tool NER2 in the brat format
from medkit.io.brat import BratOutputConverter
in_path = data_dir / "mtsamplesen" / "unannotated_doc"
# reload raw documents
final_docs = TextDocument.from_dir(
path=Path(in_path),
pattern='[A-Z0-9].txt',
encoding='utf-8',
)
# simplified pipeline, with only the best NER tool (NER2)
pipeline2 = Pipeline(
steps=[
PipelineStep(
sentence_tokenizer,
input_keys=["full_text"],
output_keys=["sentence"],
),
PipelineStep(
pii_detector,
input_keys=["sentence"],
output_keys=["sentence_"],
),
PipelineStep(
bert_matcher,
input_keys=["sentence_"],
output_keys=["ner2_drug"],
),
],
input_keys=["full_text"],
output_keys=["ner2_drug"],
)
doc_pipeline2 = DocPipeline(pipeline=pipeline2)
doc_pipeline2.run(final_docs)
# filter annotations to keep only drug annotations
# sensitive information can also be removed here
output_docs = [
TextDocument(text=doc.text, anns=doc.anns.get(label="m"))
for doc in final_docs
]
# Define Output Converter with default params,
# transfer all annotations and attributes
brat_output_converter = BratOutputConverter()
out_path = data_dir / "mtsamplesen" / "ner2_out"
# save the annotation with the best tool (considering F1 only) in `out_path`
brat_output_converter.save(
output_docs,
dir_path=out_path,
doc_names=["ner2_6", "ner2_7"],
)
Annotations of the discharge summary 6.txt, displayed with Brat

Annotations of the discharge summary 7.txt (partial view), displayed with Brat